Mayfly Phylogenetics - Current Projects
This research aims to elucidate the phylogenetic relationships of the mayfly family Coloburiscidae (Ephemeroptera) - the spinose gilled mayflies. This family is comprised of three genera: Murphyella, Coloburiscoides, and Coloburiscus. A unique characteristic of this family is that they demonstrate Gondwanan distribution being found in New Zealand, Australia, and Southern South America. In past studies, combined morphological and molecular data have questioned the family’s monophyly. The molecular data that has been used mostly comes from five “traditional” genes used in insect molecular phylogenetics. We compared our newly generated phylogenomic data to these traditional genes. We used targeted capture next generation sequencing to generate over 400 exons from the mayfly genome to create a large phylogenomic dataset. Bioinformatic software was used to align the data and carry out phylogenetic tree reconstruction using maximum likelihood, Bayesian, and maximum parsimony analyses. The resulting trees support the monophyly of Coloburiscidae, confirming the hypothesis of this research.

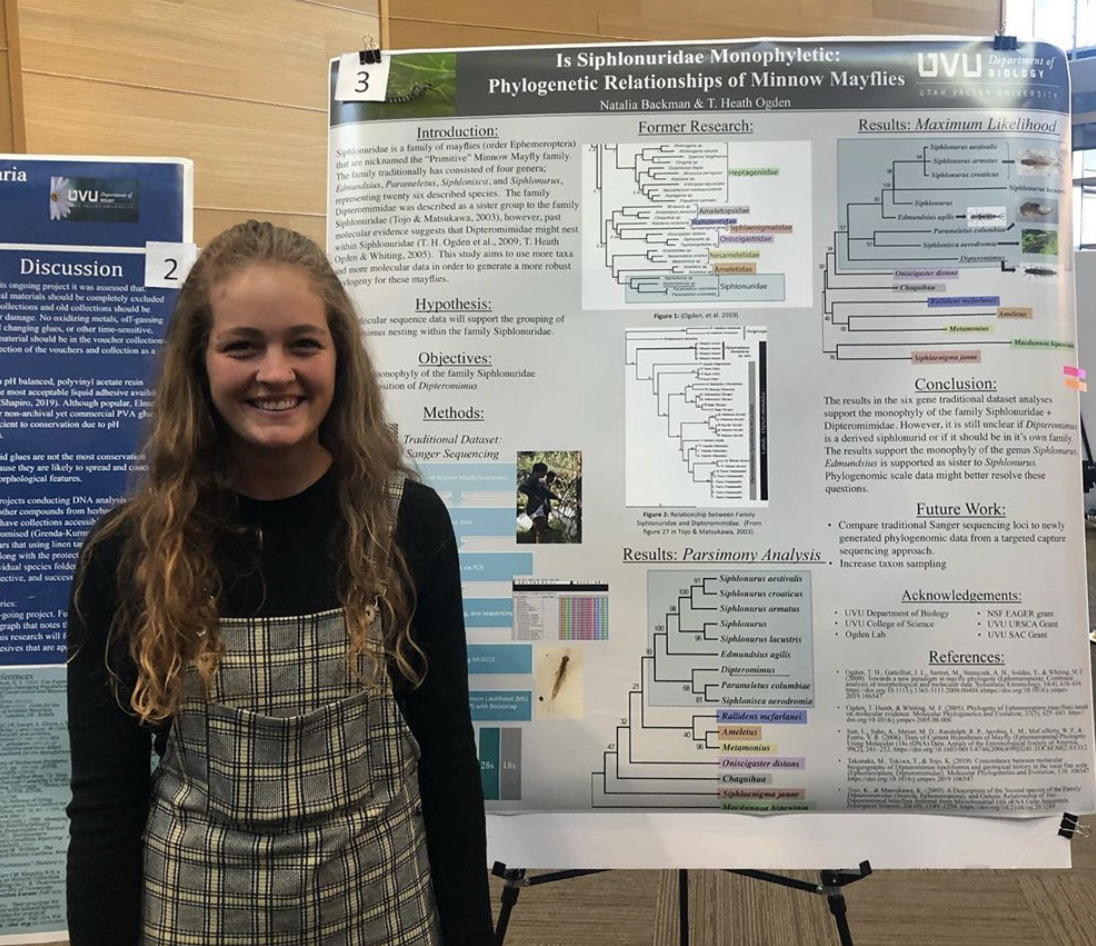
Siphlonuridae is a family of mayflies (order Ephemeroptera) that are nicknamed the “Primitive” Minnow Mayfly family. The family traditionally has consisted of four genera; Edmundsius, Parameletus, Siphlonisca, and Siphlonurus, representing twenty six described species. The family Dipteromimidae was described as a sister group to the family Siphlonuridae (Tojo & Matsukawa, 2003), however, past molecular evidence suggests that Dipteromimidae might nest within Siphlonuridae (T. H. Ogden et al., 2009; T. Heath Ogden & Whiting, 2005). This study aims to use more taxa and more molecular data in order to generate a more robust phylogeny for these mayflies.
General Entomology
Bioinformatics
We are interested in using transcriptome data, generated with next generation sequencing technology, to investigate the evolutionary trends of specific genes and their associated expression in mayflies. We generated an additional transcriptome for mayflies. RNA was extracted from a freshly frozen specimen preserved in RNAlater® (Ambion) using TRIzol® Reagent (Ambion) and cDNA libraries were prepared from mRNA. RNA-seq data was generated using a paired-end protocol (PE100) on Illumina HiSeq2000 with an expected 60 million reads. In order to effectively investigate the large amount of sequences, we created a bioinformatics workflow to analyze the newly generated transcriptome data along with previous data for mayflies. The workflow consists of these main steps: Trinity (Assembling the transcripts), Transdecoder (Identifying candidate coding regions), HMMER (Searching biological sequence databases for homologous sequences). We tested the workflow looking at opsin genes.

Pedagogical Research